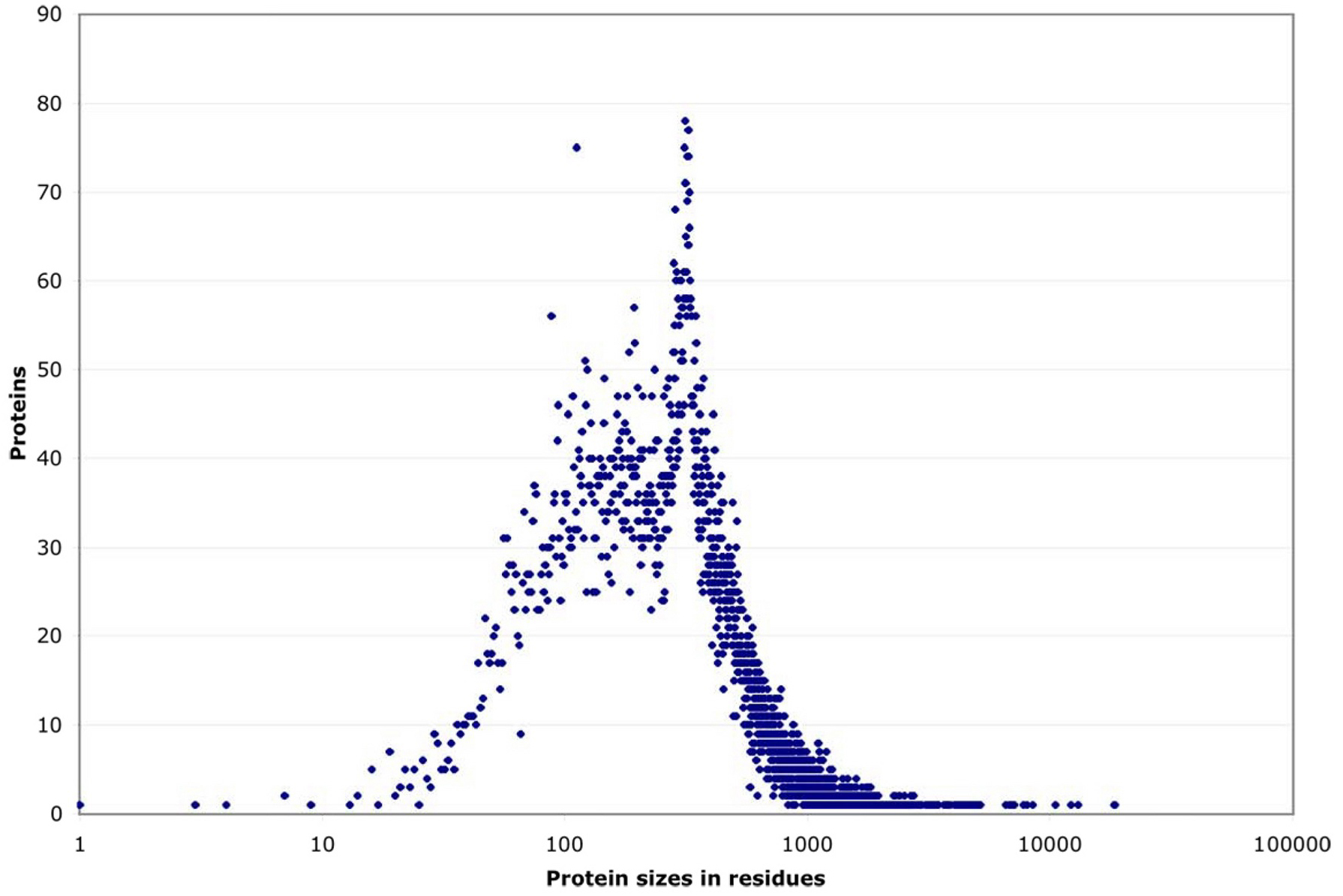
Source: wormbook.org. (Click image to enlarge.)
It would be interesting to see a similar graph for E. coli (and other organisms). I suspect the spike at ~300 residues is unique to C. elegans. If you "back out" the spike, the distribution looks fairly Gaussian.
What would be interesting is to indicate certain classes of proteins in red (or some other distinguishing color), such as proteins that require special help in folding. Naïvely, one would expect larger proteins to require more assistance with folding. But it could be more complicated than that: What if proteins that require some sort of special "folding assistance" tend to clump up at particular spots in the distribution?
